Photo by Shahadat Rahman on Unsplash
This is the third post of a series dedicated to anonymization. In the first one the fundamentals of the de-identification problem were explained; while, in the second one the methodology adopted at Konplik Health was described.
This post includes a detailed overview of the implementation of an anonymization system at Konplik Health. The starting point is a collection of anonymized EHRs available as part of the ‘2006 De-identification and Smoking Status Challenge’, organized as part of the I2B2 initiative. I2B2 is a member-driven non-profit foundation developing an open-source/open-data community around the i2b2, tranSMART and OpenBEL translational research platforms. The NLP Datasets, Challenges and Shared tasks are now hosted by the Department of Biomedical Informatics (DBMI) at Harvard Medical School as ‘n2c2: National NLP Clinical Challenges’.
An example of the result obtained from Konplik’s anonymization process when applied to one of the clinical documents included in this collection is shown below, (the collection can be accessed under request through the DBMI Data portal).

The purpose of the task would be to identify any instances of PHI replacing them with a sequence of non-representative characters (i.e, ‘XXX’) or with a label indicating the semantic type for the entity (PERSON, DATE, ORGANIZATION, …) and linking different mentions to the same entity appearing in the input text.
We applied the methodology described in our previous post when creating this anonymization model.
1. Data Preparation
The input data collection is provided as an XML file containing all the clinical reports used for training separated by XML tags and the corresponding report identifier. Firstly, this XML was broken down into numerous files, one per report and special characters were transformed. As this is a training collection, it comes with the PHI elements identified; so, as a second step, the corresponding labels were extracted and stored for each document. This training collection contains 669 clinical reports.
Data preparation is the task that consumes the most effort in any artificial intelligence project. It is not only a data formatting process but, it also allows you to gain a deep knowledge of the characteristics, statistics and relations across the data.
There is also a test set containing 889 reports that has been processed accordingly and used to validate the model obtained.
In some business cases at Konplik Health, when enough training data is available, a deep learning model has been used as the base model which is fine-tuned afterwards using a set of rules.
2. Base anonymization model performance analysis
The next step according to our methodology is to measure precision obtained with the basic model, that is, a generic model for PHI detection available at the company. MeaningCloud, Konplik’s sister company, has been working for more than 20 years in the development of Natural Language Processing tools. One of the APIs provided by MeaningCloud is the Topic Extraction API, which is able to recognize entities that are mentioned in a text. These entities can come in many different shapes and sizes, from names of places, organizations or people to money or time expressions, to name just a few. Identification capabilities of the Topic Extraction API go far beyond the needs of PHI detection. Nevertheless, it is a good starting point to build an anonymization model.
The output generated by the Topic Extraction API and the collection of labeled EHRs provided by the I2B2 task organizers are then combined to create an initial version of the anonymization model which, besides using recognized entities, is able to find relations among them and other language elements through a rule-based language.
3. Customized anonymization model generation
At this point there is a baseline model to work with and a refinement iterative process begins. Konplik’s models are built around a rule-based language developed in-company. A sample of these rules is shown in the image below.


The first rule is intended to locate names of doctors and the second one to identify any patients’ professions that may be mentioned in the input text. Of course, there are many more rules in the model, approximately 140.
The language used to define the rules is extremely powerful; it does not just allow operations similar to regular expressions but also uses semantic information provided by a linguistic parsing process that supports the analysis. Furthermore, additional dictionaries can be integrated in the process to account for specialized terminology relevant to the domain where the rules have been applied.
The recognized elements are arranged into a graph structure (as you have perhaps imagined when reading the CREATE_NODE operation featured in the rules above). This allows for further analysis and facilitates the understanding of relations among the text elements recognized.
The anonymization rules are edited through a model development workbench, shown in the figure below.
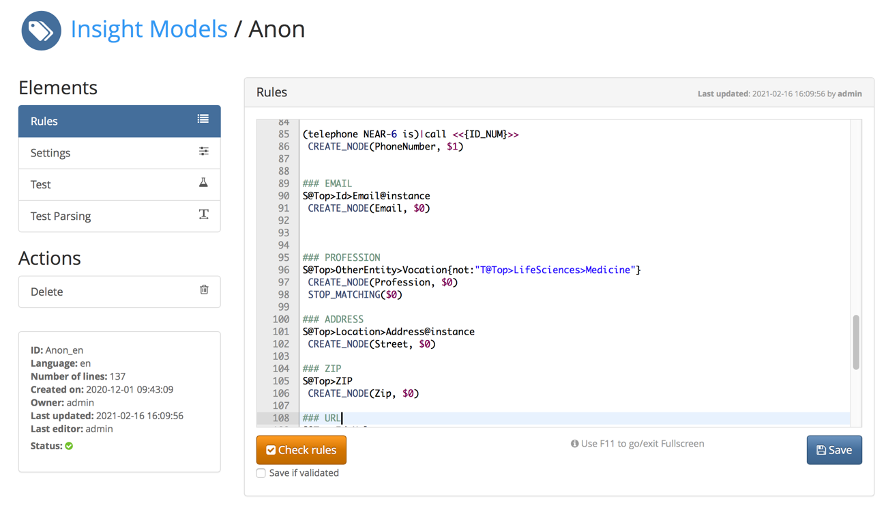
The rule’s workbench simplifies the work of Konplik’s computational linguistics team who generate, validate and maintain the anonymization model. It is the main tool that simplifies the iterative development process, which starts with the definition of the rule, that is then tested against the training input text and modified if needed. This process is repeated until all the desired elements are recognized.
4. Customized anonymization model deployment
Once the anonymization model is available, it is not enough to just measure its precision, as it is done in research challenges. The model must be deployed in a production environment and applied to new EHRs generated at the hospital or health institution daily.
The deployment process is really easy for Konplik, as a REST Application Programming Interface (API) based infrastructure is already available. Given that data privacy must be minded at all times, an on-premises deployment is advised, in a way that EHRs do not have to travel out of the hospital’s private network.
5. Model maintenance
The work does not finish when the model is deployed to production, it must be maintained, which means that the validity of the generated output must be periodically checked. It can happen that new linguistic structures are used in reports (at least, structures that were not seen during model development, as it is almost impossible to have a training collection which covers all linguistic cases) so the model needs to be updated accordingly. This is a key issue in any artificial intelligence project; as is true for data, models are alive, that is, they must be reviewed and updated if needed.
In summary, this post shows the way Konplik Health implements EHR de-identification models, introducing both the rule-based language available to define these models and the process which brings them to life.
Languages
The model described is intended for EHRs in English However, Konplik Health has also developed a model for Spanish EHRs, based on the training collections provided by the Medical Document Anonymization (MEDDOCAN) challenge organized as part of the Spanish Initiative to promote Language Technologies. Additional languages will be covered in the coming months.
Please, do not hesitate to leave any comment you may have and get in touch if you are interested in knowing how it can be applied to your particular business case.


Recent Comments